Contents
Introduction
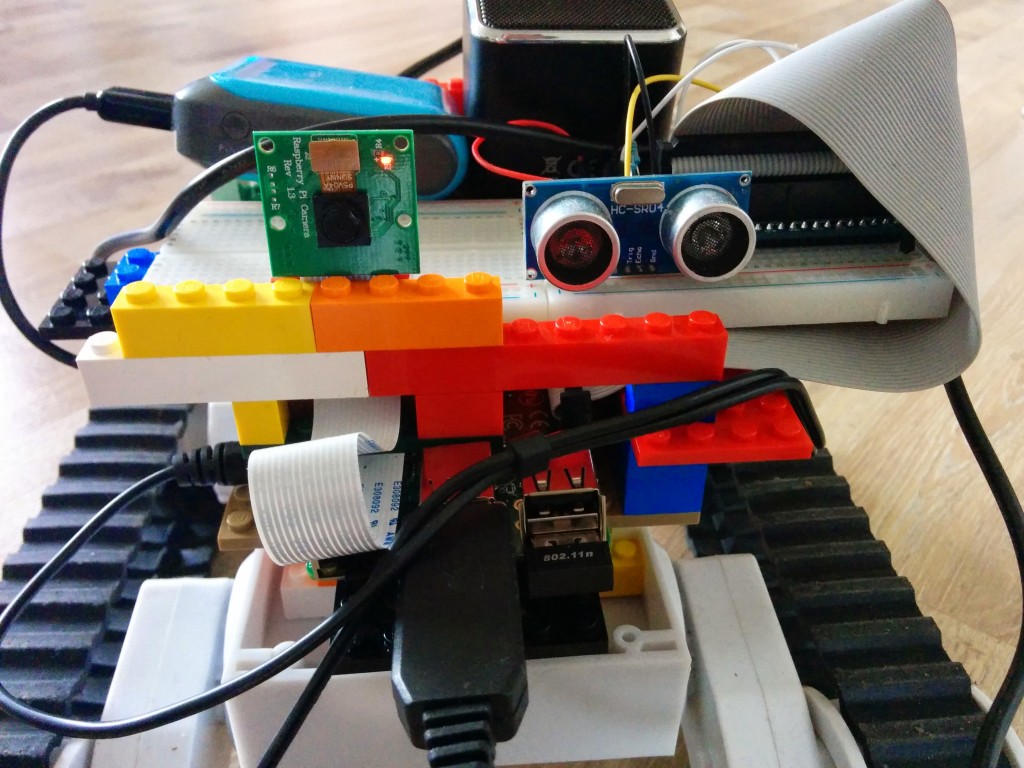
The HC-SR04 can be used to measure distance and detect obstacles in robotic projects. However, the reported range is not always accurate, small objects are mostly not detected at all and sometimes false positives occur (a short range is reported when the way ahead is free). That said, we’ll give it a try to report ranges that, for instance, to prevent a Pi Rover from running into walls. Furthermore, I’ll undertake an attempt to eliminate bogus values.
Connecting the sensor
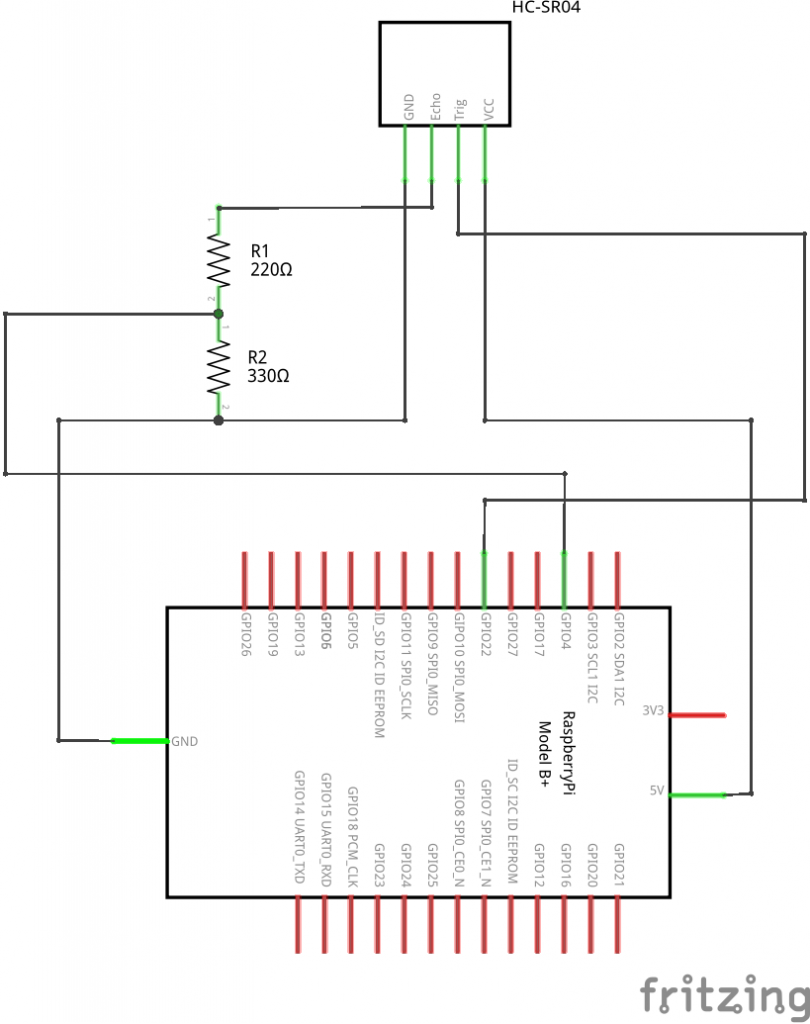
The HC-SR04 has 4 pins. Power (5 V DC) is supplied over the VCC and GND pins. A pulse is initiated on the Trig pin, which takes 3.3 V. The result is measured on the Echo pin, which delivers 5 V. So to use it with Raspberry Pi GPIO we need bring it down to around 3.3 V. We’ll use a potential divider for that.
A potential divider divides the potential over two serial resistors, as illustrated here:
The relationship between the input and output voltage is:
So with R1 = 220 Ω and R2 = 330 Ω we have an output of 3 V. That’s good enough. The GPIO pin needs a voltage of at least 2.5 V to read positive.
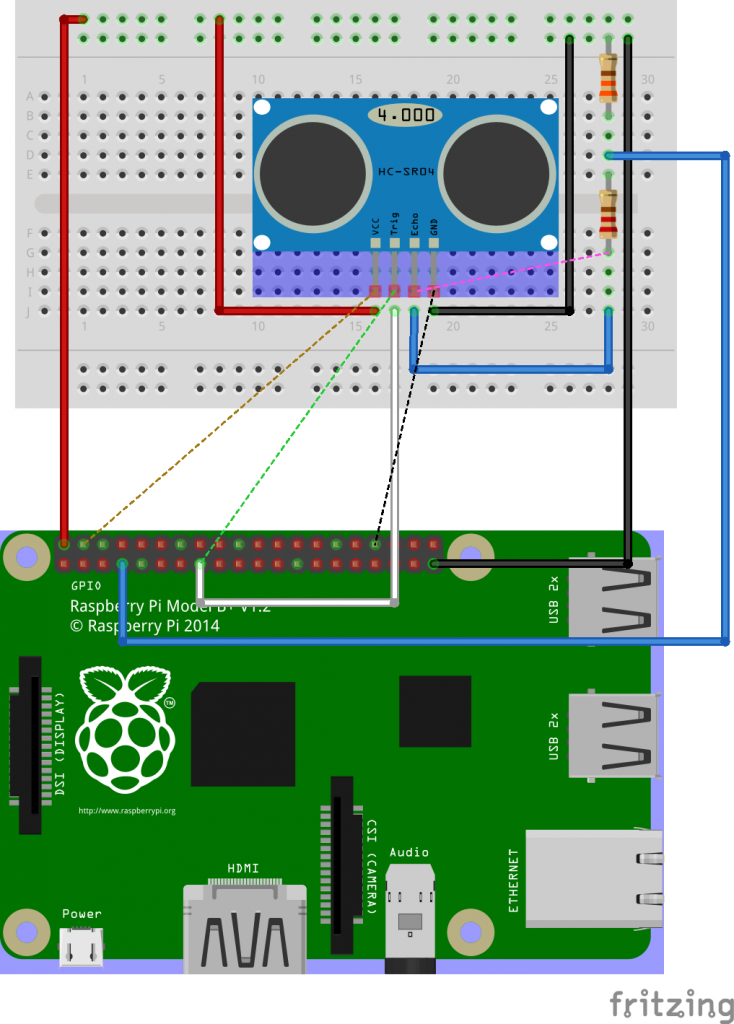
We’ll connect the trigger to GPIO22 (physical pin 15) and the echo over the potential divider to GPIO4 (physical pin 7).
Collecting data
You might expect that the delta time between the trigger and the echo reflect the distance measured. This is not quite the case. The HC-SR04 expects a pulse of at least 10 μs as a trigger to fire a burst of 8 ultrasound pulses (40 kHz). The distance corresponds to the length of the echo pulse. We multiply this value by the speed of sound in cm to find the total range in cm. Dividing by two gives the distance to the object.
Let’s collect some data in Python. We’ll use numpy for some calculations (sudo pip3 install numpy) and WiringPi to access GPIO.
#!/usr/bin/python3 import wiringpi2 import time import datetime import numpy as np ON = 1 OFF = 0 IN = 0 OUT = 1 TRIGGER = 15 ECHO = 7 PULSE = 0.00001 BURST = 25 SPEED_OF_SOUND = 34029 wiringpi2.wiringPiSetupPhys() wiringpi2.pinMode(TRIGGER, OUT) wiringpi2.pinMode(ECHO, IN) wiringpi2.digitalWrite(TRIGGER, OFF) def pulse(): wiringpi2.digitalWrite(TRIGGER, ON) time.sleep(PULSE) wiringpi2.digitalWrite(TRIGGER, OFF) starttime = time.time() stop = starttime start = starttime while wiringpi2.digitalRead(ECHO) == 0 and start < starttime + 2: start = time.time() while wiringpi2.digitalRead(ECHO) == 1 and stop < starttime + 2: stop = time.time() delta = stop - start distance = delta * SPEED_OF_SOUND return distance / 2.0 def measure_range(): values = np.zeros(BURST) for i in range(0, BURST): values[i] = pulse(); print("Measured: %1.1f" % values[i]) time.sleep(0.08) print(repr(values)) return np.mean(values); print("Range: %1.1f" % measure_range()) |
Note that a pulse in our program results in 8 ultrasound pulses in the HC-SR04. The sleep time between the (software) pulses will interfere with the measurement if it is too short, especially for longer ranges. This does not need to bother us much. First, the sensor is inaccurate above 4 m anyway. Second, 4 m and 20 m essentially mean the same thing to the Pi Rover, namely: the way ahead is free.
If we run this the output may look like the following:
Measured: 238.8
Measured: 3361.1
Measured: 239.9
Measured: 2352.1
Measured: 243.3
Measured: 241.6
Measured: 240.5
Measured: 241.0
Measured: 240.9
Measured: 215.1
Measured: 114.1
Measured: 241.5
Measured: 241.1
Measured: 242.0
Measured: 122.5
Measured: 242.0
Measured: 240.4
Measured: 241.7
Measured: 241.1
Measured: 244.5
Measured: 241.7
Measured: 118.3
Measured: 119.7
Measured: 242.2
Measured: 240.2
array([ 238.78205144, 3361.14650631, 239.85298669, 2352.13484251,
243.32135653, 241.64193535, 240.50203836, 240.99288368,
240.87118649, 215.13223135, 114.08300006, 241.48778558,
241.09429801, 241.99891376, 122.52067173, 242.01108348,
240.41685033, 241.67033136, 241.09429801, 244.51398897,
241.65410507, 118.30183589, 119.69729698, 242.2179687 ,
240.15722966])
Range: 429.9
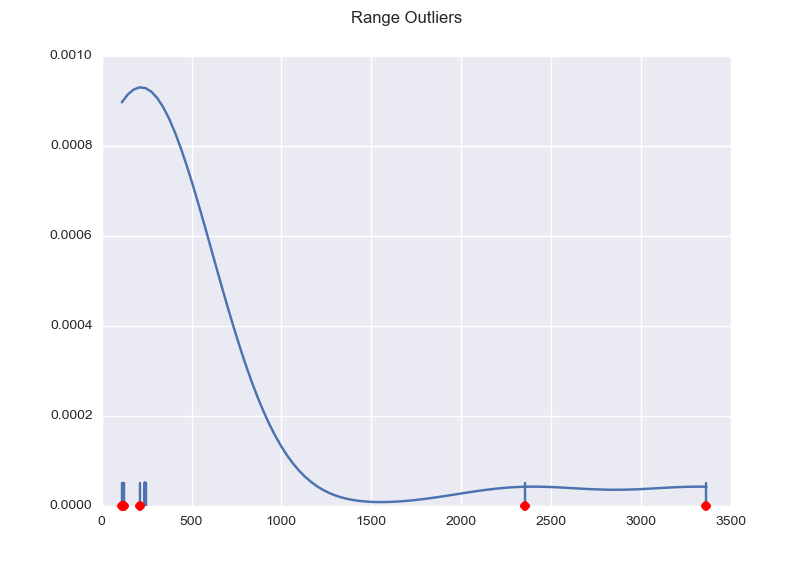
Now the range is clearly about 240 cm, but with the ultrasound scattering all over the place we get some values that are obviously not correct. So averaging the data does not appear to make much sense. We need to eliminate the outliers first.
Finding the outliers
Here’s an excellent explanation on finding outliers in Python: http://stackoverflow.com/a/22357811/959505. We’ll use the median-absolute-deviation method.
Skip to the next section if you don’t want to experiment with this. I’m doing this in PyCharm on a Linux PC and I needed to compile a lot of libraries (and even install a Fortran compiler) to get SciPy working. You probably don’t want to do this on a Raspberry Pi.
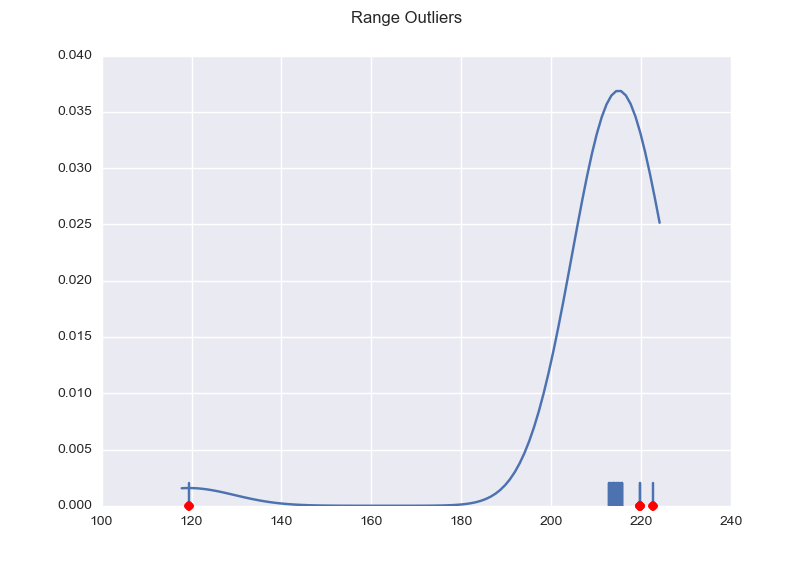
import numpy as np import matplotlib.pyplot as plt import seaborn as sns def main(): x = np.array([ 238.78205144, 3361.14650631, 239.85298669, 2352.13484251, 243.32135653, 241.64193535, 240.50203836, 240.99288368, 240.87118649, 215.13223135, 114.08300006, 241.48778558, 241.09429801, 241.99891376, 122.52067173, 242.01108348, 240.41685033, 241.67033136, 241.09429801, 244.51398897, 241.65410507, 118.30183589, 119.69729698, 242.2179687 , 240.15722966]) plot(x) plt.show() def mad_based_outlier(points, thresh=3.5): if len(points.shape) == 1: points = points[:,None] median = np.median(points, axis=0) diff = np.sum((points - median)**2, axis=-1) diff = np.sqrt(diff) med_abs_deviation = np.median(diff) modified_z_score = 0.6745 * diff / med_abs_deviation return modified_z_score > thresh def plot(x): fig, ax = plt.subplots() sns.distplot(x, ax=ax, rug=True, hist=False) outliers = x[mad_based_outlier(x)] ax.plot(outliers, np.zeros_like(outliers), 'ro', clip_on=False) fig.suptitle('Range Outliers') main() |
This results in the following graph, with the outliers marked red and the useful values nicely clustered.
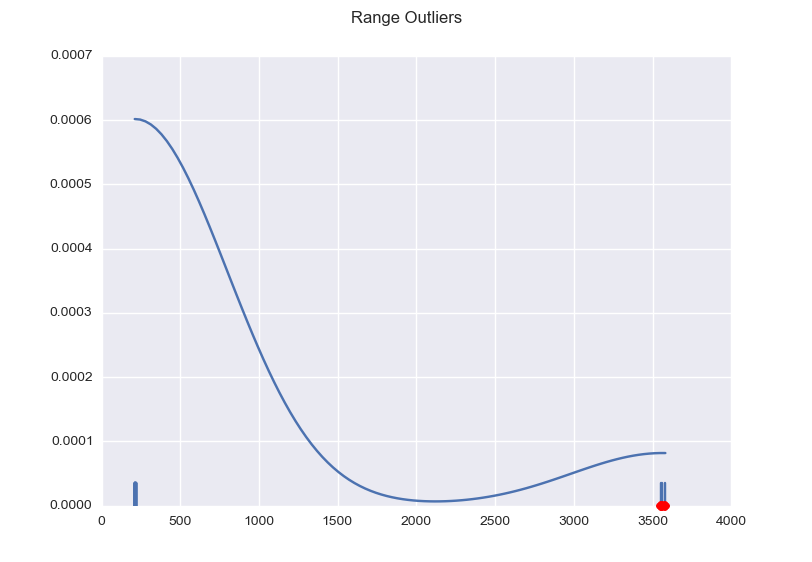
Here are two more examples to give you an idea of the possible dispersion.
Filtering the data
So what we need to do is pass our data through the mad_based_outlier function to filter out the rubbish. In this final example the Pi Rover sends a burst of 5 pulses (again, 5 x 8 = 40 ultrasound pulses), measures the results, cleans up the data and pauses 200 ms before repeating the cycle. Experiment with these values to obtain optimal results. When an obstacle is detected the Pi Rover reports this and stops its engines.
!/usr/bin/python3 import wiringpi2 import time import datetime import numpy as np from subprocess import call from subprocess import Popen ON = 1 OFF = 0 IN = 0 OUT = 1 TRIGGER = 15 ECHO = 7 PULSE = 0.00001 BURST = 5 SPEED_OF_SOUND = 34029 wiringpi2.wiringPiSetupPhys() wiringpi2.pinMode(TRIGGER,OUT) wiringpi2.pinMode(ECHO,IN) wiringpi2.digitalWrite(TRIGGER,OFF) obstacle_reported = False reported_range = 0 def pulse(): wiringpi2.digitalWrite(TRIGGER,ON) time.sleep(PULSE) wiringpi2.digitalWrite(TRIGGER,OFF) starttime = time.time() stop = starttime start = starttime while wiringpi2.digitalRead(ECHO) == 0 and start < starttime + 2: start = time.time() while wiringpi2.digitalRead(ECHO) == 1 and stop < starttime + 2: stop = time.time() delta = stop - start distance = delta * SPEED_OF_SOUND return distance / 2.0 def mad_based_outlier(points, thresh=3.5): if len(points.shape) == 1: points = points[:,None] median = np.median(points, axis=0) diff = np.sum((points - median)**2, axis=-1) diff = np.sqrt(diff) med_abs_deviation = np.median(diff) modified_z_score = 0.6745 * diff / med_abs_deviation return modified_z_score > thresh def measure_range(): values = np.zeros(BURST) for i in range(0, BURST): values[i] = pulse(); print("Measured: %1.1f" % values[i]) time.sleep(0.01) filtered = values[~mad_based_outlier(values)] print(repr(values)) print(filtered) return np.mean(filtered); while True: print(obstacle_reported) range_result = measure_range() if range_result < 50 and not obstacle_reported: # shell script calling espeak Popen(['./say', 'obstacle ahead']) # python program to send stop commands to the motor controller call(['./dcmotorstop.py', '']) obstacle_reported = True reported_range = range_result elif range_result > (reported_range + 10): obstacle_reported = False time.sleep(0.2) |